Genetic dissection of behavioral and neurogenomic responses to acute ethanol
Abstract
Individual differences in initial sensitivity to ethanol are strongly related to the heritable risk of alcoholism in humans. To elucidate key molecular networks that modulate ethanol sensitivity we performed a systems genetics analysis of ethanol-responsive gene expression in brain regions of the mesocorticolimbic reward circuit (prefrontal cortex, nucleus accumbens and ventral midbrain) across the BXD recombinant inbred panel, a highly diverse family of isogenic mouse strains before and after treatment with ethanol.
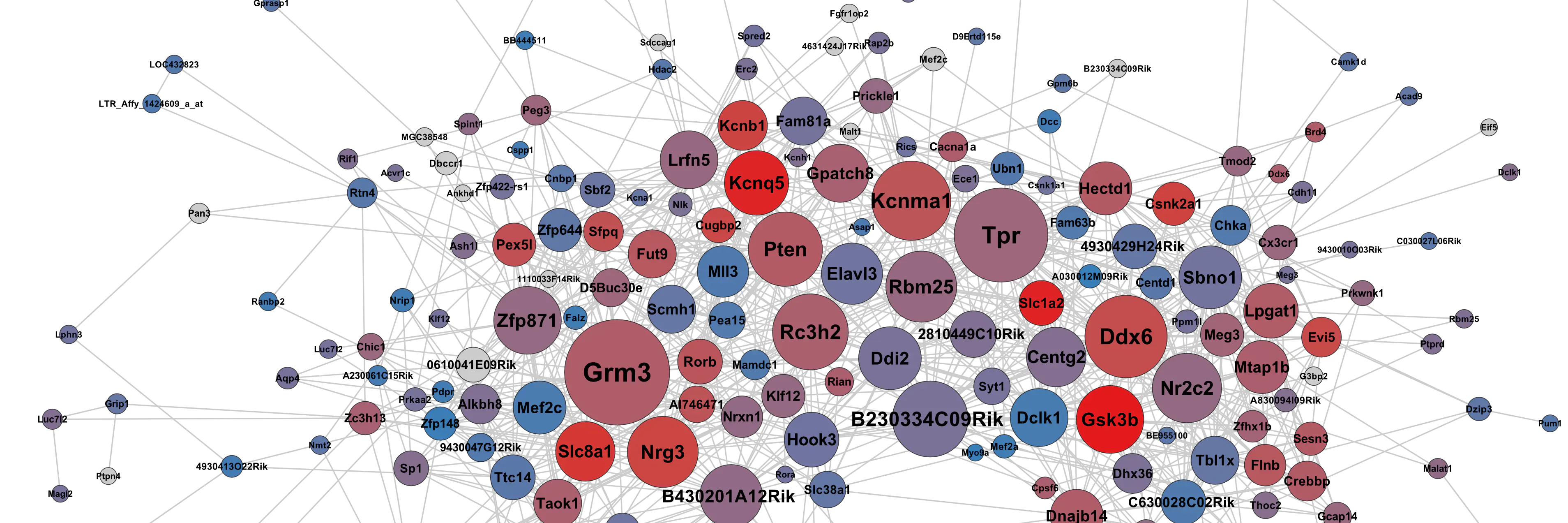
Acute ethanol altered the expression of ~2,750 genes in one or more regions and 400 transcripts were jointly modulated in all three. Ethanol-responsive gene networks were extracted with a powerful graph theoretical method that efficiently summarized ethanol’s effects. These networks correlated with acute behavioral responses to ethanol and other drugs of abuse. As predicted, networks were heavily populated by genes controlling synaptic transmission and neuroplasticity.
Several of the most densely interconnected network hubs, including Kcnma1 and Gsk3β, are known to influence behavioral or physiological responses to ethanol, validating our overall approach. Other major hub genes like Grm3 and Nrg3 represent novel targets of ethanol effects. Networks were under strong genetic control by variants that we mapped to a small number of chromosomal loci. Using a novel combination of genetic, bioinformatic and network-based approaches, we identified high priority cis-regulatory candidate genes, including Scn1β, Gria1, Sncβ and Nell2.
The ethanol-responsive gene networks identified here represent a previously uncharacterized intermediate phenotype between DNA variation and ethanol sensitivity in mice. Networks involved in synaptic transmission were strongly regulated by ethanol and could contribute to behavioral plasticity seen with chronic ethanol. Our novel finding that hub genes and a small number of loci exert major influence over the ethanol response of gene networks could have important implications for future studies regarding the mechanisms and treatment of alcohol use disorders.
Supplementary tables
-
Table S1 Lists of genes found to be significantly responsive to acute ethanol across the BXD RI panel in prefrontal cortex (PFC), nucleus accumbens (NAc) and ventral midbrain (VMB).
-
Table S2 Functional over-representation analysis of ethanol responsive genes in Table S1.
-
Table S3 Lists of genes found to be significantly responsive to acute ethanol across LXS RI panel in PFC.
-
Table S4 List of genes that belong to basal or ethanol responsive co-expression networks in PFC of BXD RI panel. Networks were generated by paraclique analysis of RMA (saline/ethanol treatment) or S-score (saline vs ethanol) expression data. Degree of connectivity and betweenness centrality measures are provided for each probe-set.
-
Table S5 Overlap of paraclique networks provided in Table S4.
-
Table S6 Peak eQTL results for all members of the basal and ethanol responsive paraclique networks at least one suggestive eQTL (genome-wide p-value < 0.63).
-
Table S7 Ranking results of positional candidate genes underlying each of the major ethanol-responsive network’s eQTL trans-bands.
-
Table S8 Functional over-representation analysis of PFC S-score networks provided in Table S4.
-
Table S9 List of Affymetrix GeneChip Mouse 430 2.0 probe-sets that overlap one or more B6/D2 polymorphisms.
R source code
-
fisher_sscoreUse Fisher’s combined probability test to analyze S-score expression data. -
ggAffy_ProbePlotggplot2 wrapper to visualize probe-level intensity data for multiple probe-sets from an AffyBatch object.